Rhinovirus is the most common virus in flu infections, while the pneumococcus is responsible for many cases of meningitis. Covid causes colds in temperate seasons and the flu in winter. Besides the disease for which they were named, germs can cause other diseases and different diseases in different seasons. Moreover, co-infections should outnumber infections from a single germ, considering that many microbes are still not testable. Scientific evidence disproves the assumption that historical pandemics are due to a single germ and that identifying and combating it is enough to stop or slow down an epidemic.
The Pandemic Plan
Throughout history, the world has faced various pandemics that have resulted in significant human losses. The Black Plague in 1350 reduced Europe’s population from 80 to 30 million; smallpox, endemic in Europe and Asia, decimated the Americas’ population after its discovery. The Spanish Flu in 1918 resulted in 20 million deaths. Penicillin was discovered in 1928 but commercialized in 1943, leaving the viral front open, with only one weapon against it: the vaccine. Invented in 1796 for smallpox, it became mandatory in Italy in 1888. Enforced in many countries, it quickly showed results: in May 1979, WHO declared the virus eradicated.
The World Health Organization (WHO) was established in 1948 to manage infectious diseases, becoming the reference point for pandemic surveillance and preparedness. The WHO monitors viruses and develops a global epidemiological and viral detection plan, preparing an annual vaccine (1) to mitigate the impact of the flu. In 2003, the A/H5N1 avian flu virus, endemic in far eastern birds, also caused severe infections in humans. This led to the Global Influenza Preparedness Plan, known as the Pandemic Plan (PP), presented in 2005. It recommended (2) “all countries to develop and continuously update a PP following agreed guidelines” and to organize an Epidemiological and Virological Surveillance System (EVSS).
Epidemiological and Virological Surveillance in Italy
In Italy, the EVSS for influenza was launched in 1999-2000 by the Higher Institute of Health (ISS): InfluNet involves general practitioners, sentinel pediatricians – about 1000 – and the ASL Laboratories, Regions, and the National Center for Influenza, to monitor the start, duration, and intensity of the flu, as well as the circulation of various viral strains. The sentinels report cases of flu-like syndrome (ILI) and, since 2010, also ARI (Acute Respiratory Infection) from the 42nd week of the year to the 17th of the following year. The criteria for diagnosing an ILI are “at least one of the following general symptoms: fever or mild fever, malaise/exhaustion, headache, muscle pain; at least one of the following respiratory symptoms: cough, sore throat, labored breathing.” The criteria for determining whether a virus can cause a pandemic (3), indicated by the WHO and adapted by each country (4), are mainly based on contagiosity.
Contagiosity and Relevance of Asymptomatic Infections
Measuring a virus’s contagiosity is essential to assess its impact; it’s measured by the Rt but only on symptomatic cases, as ISS explains (5): “Why do we calculate Rt only for symptomatic cases? The detection of asymptomatic infections depends a lot on the ability to perform screenings. The result is that a greater or smaller increase in asymptomatic cases found does not depend on the virus’s transmissibility but on the number of tests performed.” However, surveillance based solely on symptomatic cases may not be enough to detect asymptomatic infections and individuals in the latency period. For Covid, in Italy, up to December ’21, symptomatic cases and their contacts were tested, but from the beginning of ’22, the tests were liberalized; figure 1 shows the incidence of infections before and after, separated by a black line.
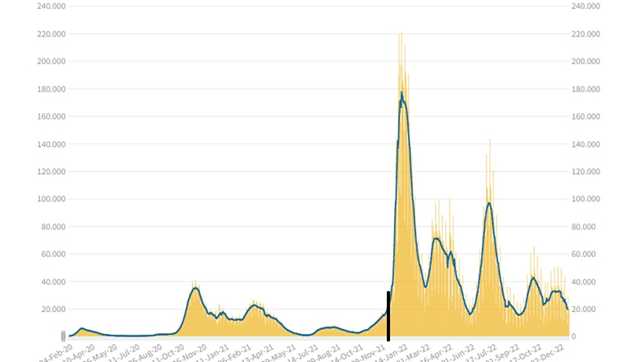
The expansion of testing has allowed for the identification of asymptomatic and presymptomatic cases, revealing that the virus’s contagiosity was higher 2-3 days before and after symptom onset (6). Therefore, it has become important to test not only symptomatic cases but also asymptomatic ones to break the transmission chain.
Co-infections
During the H1N1 pandemic period (from 11-06-2009 to 06-08-2010), a virological study (7) was underway in Glasgow, which tested 44,230 respiratory infection cases contracted from 2005 to 2013 with a multiplex test that identified 11 viruses. The results are as follows:
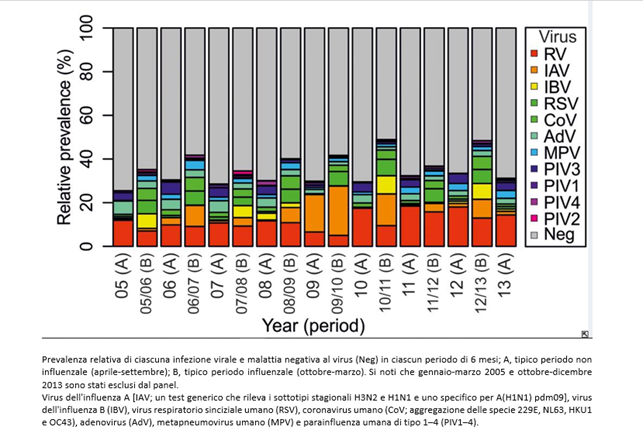
The flu is caused by a set of different viruses, most of which are not included in the tests (in gray). For simplicity, we will use the term “unknown”, but they can be microbes still without a specific test or genuinely unknown. The study, only revealing co-infections among the tested germs, significantly reduces the possibility of highlighting co-infections.
With multiplex tests, patients with respiratory infection symptoms (ARI) who tested negative for Covid19 in the 2020-21 season were also investigated in a study conducted during the Covid pandemic (8).
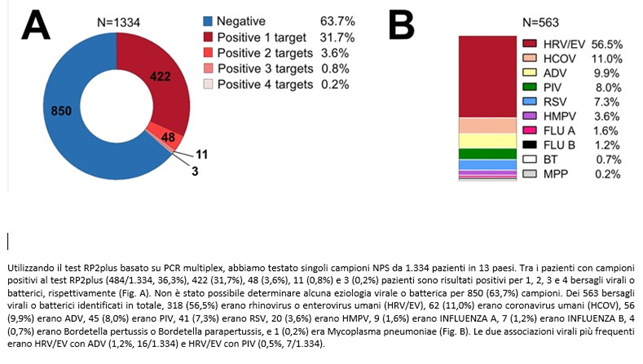
The multiplex, even including 18 viruses and 4 bacteria, found 63.7% of patients negative for the tested germs, while in many cases co-infections were detected. Rhinovirus turned out to be the most frequent virus, while FLU A-B (properly called influenza viruses) were present in only 2.8% of patients, thanks to the vaccine. In both studies, unknown germs are much more numerous than known ones; among these two groups, co-infections cannot be detected.
Studies conducted during the H1N1 and Covid-19 pandemics have detected many respiratory viruses based on flu symptoms. Is the assumption of one pandemic – one germ still tenable?
Vaccines
In clinical practice, when faced with cold symptoms, the diagnosis remains a cold, presumably a rhinovirus. Similarly, to prevent the flu, one gets vaccinated against the influenza virus, which, in the 2022-23 season in Italy, was found in 21.8% of flu patients. The flu vaccine is only suitable for 1 in 5 people! There is no data on the vaccine’s effectiveness in Italy because: “The current InfluNet/RespiVirNet data collection system was originally designed to obtain information related to influenza viruses. The detection of other respiratory viruses in the samples taken, which began in November 2022, is not currently carried out uniformly in all the laboratories participating in the network. Therefore, small distortions in calculation may occur…” (my emphasis and italics). Plus, some weeks are accompanied by the warning “It should be noted that the observed incidence in some regions is strongly influenced by the small number of doctors and pediatricians who have sent their data so far.” (original emphasis).
As the odds ratio – and consequently the effectiveness – is calculated based on the percentage of vaccinated flu patients, and precise estimates of the incidence of influenza viruses per se cannot be obtained, it is not appropriate to calculate it. In contrast, in the USA, vaccine effectiveness has been measured since the 2004-2005 season. In the 2021-22 season, it was 36%.
Should we conclude that the vaccine has low efficacy? On the contrary: in the 20-21 season (the year of the lockdown) among the samples examined by EVSS-Italy, there was no positive result for the influenza virus. The world didn’t fare differently: out of a total of 682,485 samples, 791 were positive. The vaccine works, but there was still the flu due to other germs, even if it was attenuated by pandemic measures. If for the flu, the WHO can identify the most dangerous variants in that season, we have higher effectiveness: in the 2017-18 and 2018-19 seasons in the USA, it was respectively 36% and 47%. The vaccine is a remedy targeted at a specific germ and may include various strains of the same, but the result is always limited to that microbe.
Seasonality
The EVSS assumes that the flu has its seasonality, so during the first 2 years of the pandemic (19-20 and 20-21), only suspected cases with flu-like symptoms were tested, and during the two summer seasons, infections plummeted. By applying these rules of engagement, Covid’s seasonality appeared very similar to that of the flu. From January 2022, with the liberalization of swabs, the situation, from the beginning of June to the beginning of September of 2020 vs. 2022, appeared as follows:
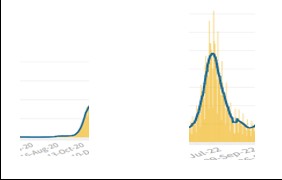
It emerged from previous studies and was also confirmed by a recent study published in Nature, that Covid-19 does not follow a fixed seasonal model and presents in different seasons with different symptoms. During the first two years, the approach of testing only patients with flu symptoms meant that the virus spread during the summer with symptoms more like a cold, thus avoiding being recognized by the tests. Therefore, Covid-19 was not identified in September but only in November when the symptoms changed with the arrival of a new season, and the virus reappeared with flu symptoms. At that point, patients were tested for Covid-19.
Mortality
The final death toll from COVID-19 is almost 7 million worldwide, compared to an annual flu death toll that is generally lower. As explained by the Higher Institute of Health on the need to combat the flu: “Acute respiratory infections caused by flu viruses can be mild, severe, and can even cause death in at-risk subjects such as the elderly and children. It is estimated that annual outbreaks cause 3 to 5 million severe cases of flu and between 290,000 and 650,000 deaths worldwide.”
The COVID-19 mortality curve by age in Italy is completely different from the previous ones and only grows with age:
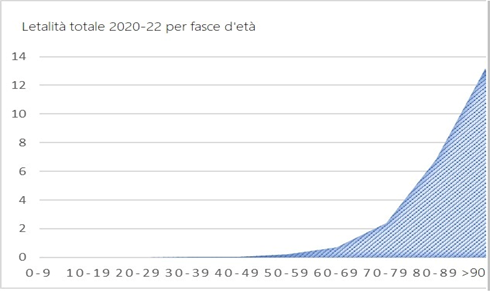
The distribution of COVID-19 victims is age-related, just like deaths from all causes:
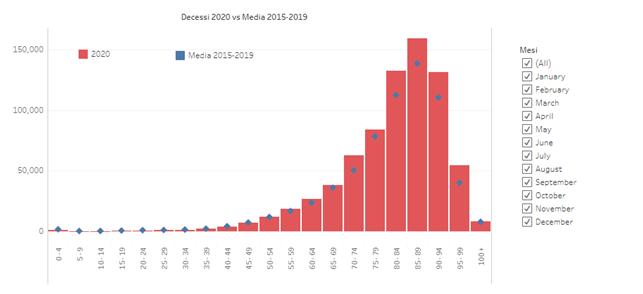
The graph shows deaths from all causes divided by age decade in the first year of COVID compared to the average of the previous 5 years: while there is no increase in mortality up to the age of 70, from 70 to 100 years there is a clear increase. The clear and continuous discrepancy is only after the age of 80.
So, what happened when there was no specific test? The last pandemics were as follows: “The flu mortality curve by age, which we know over a span of about 150 years, has always had a U-shape, with higher mortality among the very young and the elderly. However, the 1918 [Spanish flu] mortality curve was an incomplete W, similar to the U-shape, but with an added peak in middle-aged adults between 25 and 44.”
During the H1N1 pandemic, the West of Scotland Specialist Virology Centre (WoSSVC) using clinical (fever or fever history and flu-like illness or severe potentially lethal disease indicative of an infectious process) and epidemiological criteria (disease onset within 7 days of travel to an area experiencing prolonged H1N1/2009 transmission) tested 16,264 patients for H1N1/2009. Of these, only 1,516 tested positive (9%). Therefore, the study, designed specifically to detect the correlation between symptoms and diagnosis, concluded that symptom-based diagnosis was wrong in 91% of cases.
Applying this data to historical pandemics, we should conclude that only one in ten diagnoses was correct.
The only virus that would seem to be eliminated by the vaccine is emblematic, smallpox. How could a vaccine, mandatory only in a few countries and unknown in much of the world, have destroyed a virus globally? Have we eliminated a virus – a living being – or a disease – a concept that changes over time?
Considering a sample period during COVID, if 15.2 out of 100 patients with flu symptoms test positive, it is possible to assume that many of the 85 negative patients would have been mistakenly classified as positive in the absence of a specific test.
Basing statistics on symptoms and not on the specific test, all deaths showing flu signs would be attributed to the pandemic, rather than being counted as deaths due to other causes.
In the past, attributing all deaths with flu-like symptoms to the pandemic germ led to the belief that all vulnerable subjects, including children and pregnant women, were at risk during the pandemic. However, the mortality, lethality, and morbidity curves of previous pandemics differ from the current one because only deaths and patients confirmed by the specific test were counted for COVID-19.
In summary, the availability of a specific test allowed for more accurate diagnosis and helped distinguish deaths and cases related to the virus from those caused by other diseases. This has led to significant differences in statistics and death attribution compared to past epidemics where no specific test was available.
Life Expectancy and Mortality
In Italy, with about 60 million inhabitants, there have been 26 million infections and 190,000 deaths, 85% of them over 70 years old. In contrast, in Japan, with a population of about 125 million and an older demographic, 22 million infections and 46,000 deaths have been recorded, despite implementing a milder PP.
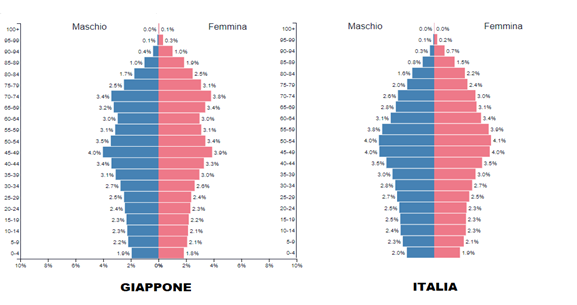
The EU nation with the highest number of COVID deaths is Bulgaria, which has an elderly population similar to other countries but a lower life expectancy. There is an even stronger correlation with health expectancy: the higher it is, the lower the contagion, morbidity, and mortality. Practically, in Italy, out of 100 people over ninety who contracted COVID, 13.2 died, while 86.8 – those with a higher life and health expectancy – survived! The emergency measures adopted by various countries have mitigated the impact of the flu complementary to these peculiarities. Therefore, given the high economic and social cost of the Pandemic Plan, a review should also be based on a cost/benefit analysis.
Aligning the Standard Model to the Evidence
Since Aristotle’s time in 400 BC, it was believed that organisms could spontaneously generate from inanimate matter. This theory, known as spontaneous generation, was accepted for over 2000 years. In the 19th century, Pasteur demonstrated through an experiment in 1861 that germs have a biological and vital nature.
On this discovery, an empirical model was built assuming that a pandemic is caused by a single germ. With various consequences: an infection like a pandemic is mainly due to a single germ.
For humans, animals, or plants, the approach started with the disease to discover the agent that caused it, and then a diagnosis was formulated based on symptoms. This approach had consequences on both taxonomy (the germs took the name of the pathology) and the etiopathogenetic model: each microbe corresponded to a disease and vice versa.
This approach allowed for symptom-based diagnoses: each germ causes a disease, and each disease is characterized by a set of symptoms that differ in some aspect from others, thus allowing differential diagnosis and therapy.
Scientific evidence in recent years has ascertained that the same germ can cause different diseases, such as, among the most common, the rhinovirus, a major factor in flu infections, or pneumococcus, responsible for many meningitis… Furthermore, the same viruses can manifest with different symptoms depending on the seasons.
Moreover, co-infections should be more than infections due to a single germ, considering that many microbes are not yet testable.
The methods to identify the germs responsible for an infection don’t help: a culture tests a limited number of bacteria and identifies the most numerous population or, better, the one that multiplies fastest in that culture. It tells us little about co-infections and amounts, and nothing about viruses. The monoplex indicates whether in a patient there is that single germ and the multiplex identifies a limited number of germs without indicating the dominant one. And we are just at the tip of the iceberg: our current ignorance does not allow us to investigate interactions in co-infections. The most investigated germ of all, COVID-19, has given clinical syndromes from mild to deadly; statistics on mortality and morbidity have clarified that life expectancy has a significant impact, but not the interactions between different microorganisms.
Scientific evidence dispels the assumption that historical epidemics are due to a single germ. And consequently, that it is enough to identify and counteract it to stop or slow down an epidemic/pandemic.
So, since these studies have been known for a long time, why hasn’t this line of research been followed? Probably because the lack of valid means to investigate this complex microbial world led us to reduce it to what we knew and could investigate. Or could it be a reluctance to implement new findings, a kind of systemic bias?
Bibliography
- EpiCentro (2023) Influenza, Vaccini antinfluenzali disponibili. Available at: https://www.epicentro.iss.it/influenza/vaccini-disponibili
- World Health Organization. (2005). WHO global influenza preparedness plan: the role of WHO and recommendations for national measures before and during pandemics. World Health Organization. Available at: https://apps.who.int/iris/handle/10665/68998
- WHO (2017) WHO guidance for surveillance during an influenza pandemic. Available at: https://apps.who.int/iris/bitstream/handle/10665/259886/9789241513333-eng.pdf
- Come funziona l’analisi del Rischio Epidemico (2020) ISS. Available at: https://www.iss.it/coronavirus/-/asset_publisher/1SRKHcCJJQ7E/content/come-funziona-lanalisi-del-rischio-epidemico.
- FAQ Sul calcolo Del Rt (2020) ISS. Available at: https://www.iss.it/covid-19-primo-piano/-/asset_publisher/yX1afjCDBkWH/content/faq-sul-calcolo-del-rt#:~:text=Per%20calcolare%20R0%20o%20Rt,criteri%20sufficientemente%20stabili%20nel%20tempo.
- Luca Ferretti, Alice Ledda, Chris Wymant et al. The timing of COVID-19 transmission. medRxiv preprint doi: https://doi.org/10.1101/2020.09.04.20188516; this version posted September 7, 2020.
- Nickbakhsh, S. et al. (2016) Extensive multiplex PCR diagnostics reveal new insights into the epidemiology of viral respiratory infections, PubMed Central (PMC). Available at: https://www-ncbi-nlm-nih-gov.translate.goog/pmc/articles/PMC7113017/?_x_tr_sl=en&_x_tr_tl=it&_x_tr_hl=it&_x_r_pto=sc.
- Duclos M;Hommel B;Allantaz F;Powell M;Posteraro B;Sanguinetti M; (2022) Multiplex PCR detection of respiratory tract infections in SARS-COV-2-negative patients admitted to the Emergency Department: An international multicenter study during the COVID-19 pandemic, Microbiology spectrum. U.S. National Library of Medicine. Available at: https://pubmed.ncbi.nlm.nih.gov/36154273/.
- Viral aetiology of influenza-like illnesses and severe acute respiratory illnesses in Morocco, September 2014 to December 2016. Abderrahman Bimouhen1,3, Zakia Regragui3, Fatima El Falaki3. https://jogh.org/wp-content/uploads/2022/07/jogh-12-04062.pdf
- Schulz, F. et al. (2020) Giant virus diversity and host interactions through global metagenomics, Nature News. Nature Publishing Group. Available at: https://www.nature.com/articles/s41586-020-1957-x.










